Contamination data of Listeria monocytogenes in smoked fish
smokedfish.RdContamination data of Listeria monocytogenes in smoked fish on the Belgian market in the period 2005 to 2007.
Usage
data(smokedfish)Format
smokedfish is a data frame with 2 columns named left and right, describing
each observed value of Listeria monocytogenes concentration (in CFU/g) as an interval.
The left column contains either NA for
left censored observations, the left bound of the interval for interval censored
observations, or the observed value for non-censored observations. The right
column contains either NA for right censored observations, the right bound of
the interval for interval censored observations, or the observed value for non-censored
observations.
Source
Busschaert, P., Geereard, A.H., Uyttendaele, M., Van Impe, J.F., 2010. Estimating distributions out of qualitative and (semi) quantitative microbiological contamination data for use in risk assessment. International Journal of Food Microbiology. 138, 260-269.
Examples
# (1) load of data
#
data(smokedfish)
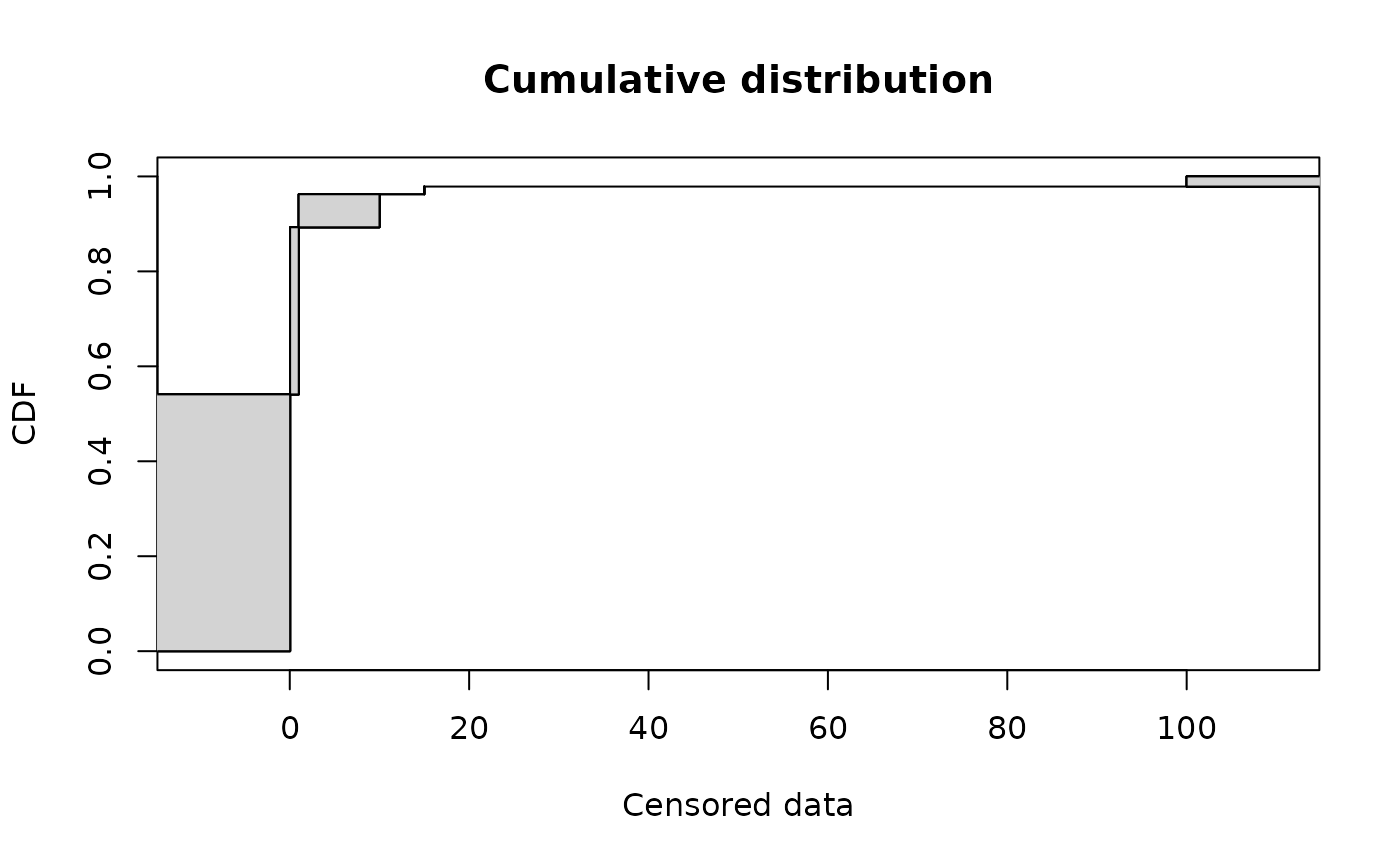
# (2) plot of data in CFU/g
#
plotdistcens(smokedfish)
 # (3) plot of transformed data in log10[CFU/g]
#
Clog10 <- log10(smokedfish)
plotdistcens(Clog10)
# (3) plot of transformed data in log10[CFU/g]
#
Clog10 <- log10(smokedfish)
plotdistcens(Clog10)
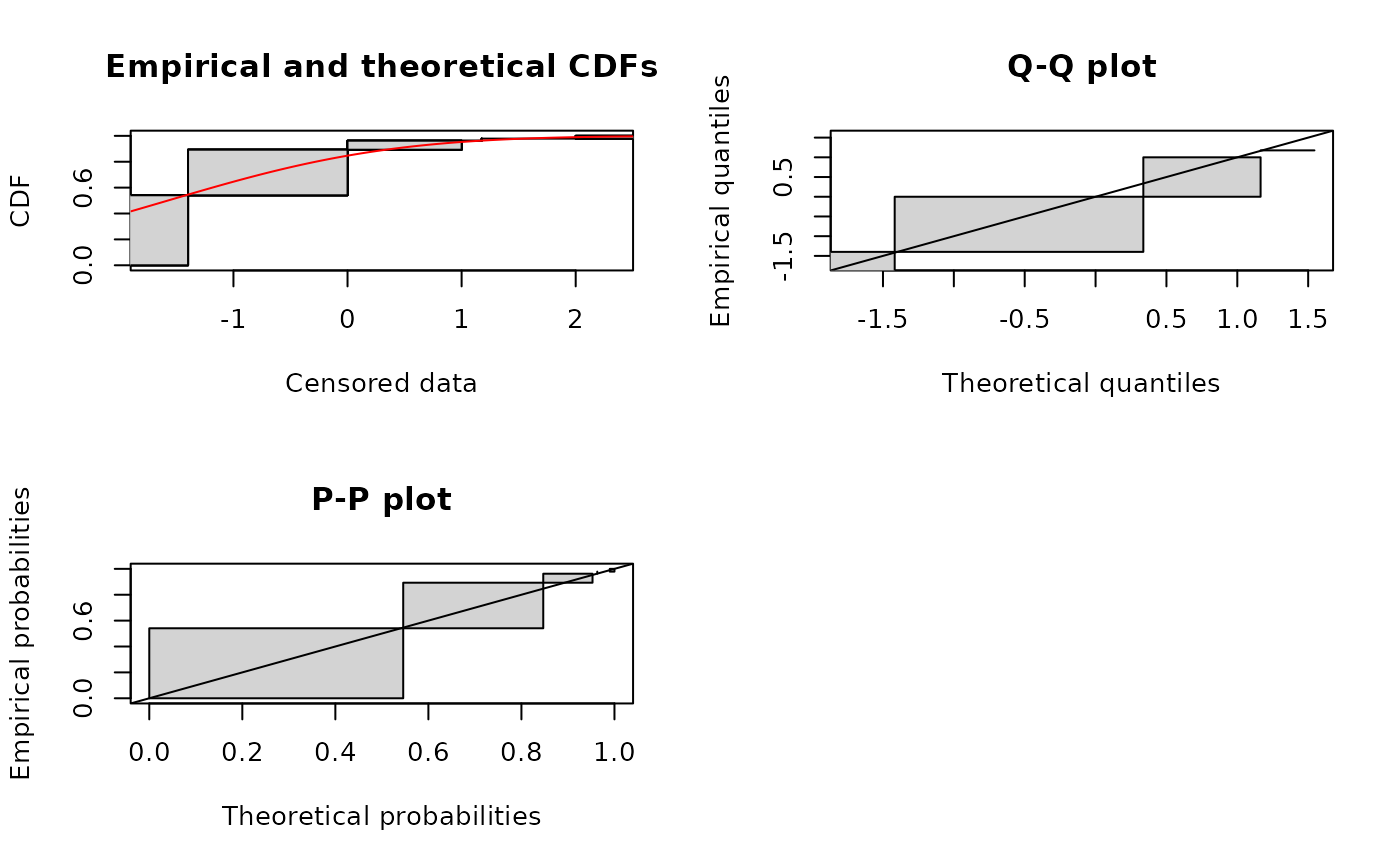
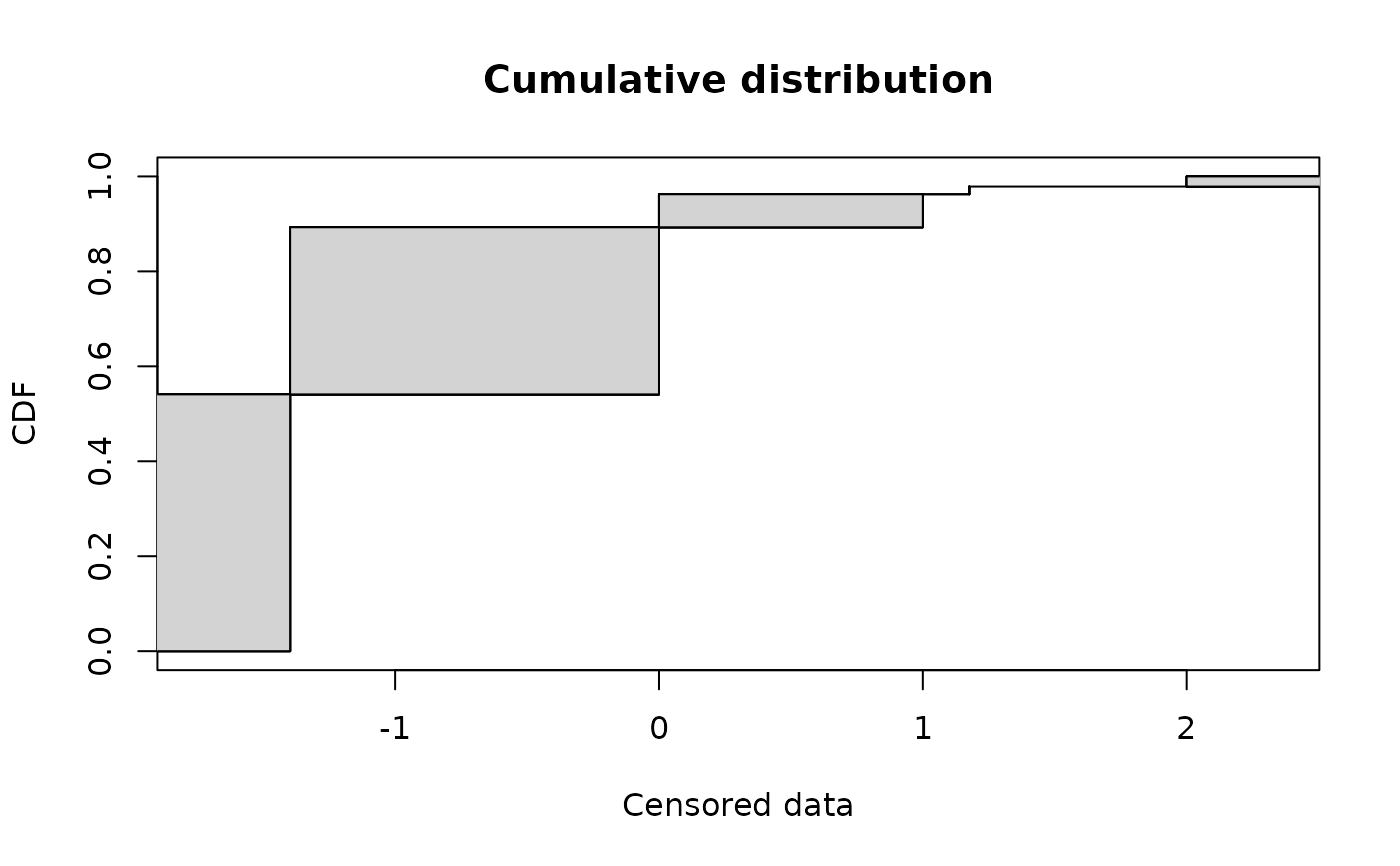 # (4) Fit of a normal distribution to data in log10[CFU/g]
#
fitlog10 <- fitdistcens(Clog10, "norm")
summary(fitlog10)
#> Fitting of the distribution ' norm ' By maximum likelihood on censored data
#> Parameters
#> estimate Std. Error
#> mean -1.575392 0.2013872
#> sd 1.539446 0.2118026
#> Loglikelihood: -87.10945 AIC: 178.2189 BIC: 183.4884
#> Correlation matrix:
#> mean sd
#> mean 1.0000000 -0.4325228
#> sd -0.4325228 1.0000000
#>
plot(fitlog10)
# (4) Fit of a normal distribution to data in log10[CFU/g]
#
fitlog10 <- fitdistcens(Clog10, "norm")
summary(fitlog10)
#> Fitting of the distribution ' norm ' By maximum likelihood on censored data
#> Parameters
#> estimate Std. Error
#> mean -1.575392 0.2013872
#> sd 1.539446 0.2118026
#> Loglikelihood: -87.10945 AIC: 178.2189 BIC: 183.4884
#> Correlation matrix:
#> mean sd
#> mean 1.0000000 -0.4325228
#> sd -0.4325228 1.0000000
#>
plot(fitlog10)